Research Groups
Our work is centred on how metal ions, such as Zn2+, Ca2+ and Co2+ are transported through the blood and how their mishandling can lead to disease.
I work on Darwinian adaptation. Natural selection explains the appearance of design in the living world, but at what level is this design expected to manifest – gene, individual, society – and what is its function?
The group’s research focusses on using Electron Paramagnetic Resonance (EPR) spectroscopy to study the microscopic structure and dynamics of (bio)molecules and materials.
We conduct research on a wide range of taxa including corals, fish, birds, aquatic invertebrates, and plants.
We are interested in how shape and form arise during animal development. During morphogenesis, cells interpret complex information and react accordingly to shape tissues and organs. Here, many cell behaviours, such as cell migration and cell shape changes, work together. Despite recent progress, it still remains mysterious how all these behaviours are regulated and coordinated.
Research is focused on identifying new treatments for Multi-Drug Resistant (MDR) human pathogens.
Novel treatments can be single therapies or combinations. Combinations can be derived from antibiotics; antibiotic resistance ‘breakers’ eg. Beta-lactamase inhibitors; efflux-pump inhibitors; or repurposed drugs.
Cells must defend their genomes against attack by selfish genetic elements such as viruses. Although CRISPR is widely known as a genome editing technology, the CRISPR system functions as an adaptive immune system in prokaryotes.
Our research combines enzymology, structural biology, chemical biology and microbiology to understand how bacteria produce and use cyclic peptides in interspecies and interkingdom warfare and/or cooperation. Our purpose is to generate novel drugs, probes, carriers, and technologies that will allow us to control and modulate bacterial populations.
In the da Silva Lab, we apply techniques of molecular biology, biochemistry, structural biology and physical organic chemistry to unravel the mechanisms of enzymatic reactions catalysed by multi-protein allosteric complexes, tRNA methyltranferases, and nucleotide hydrolases. Particular attention is given to transition-state structure, inhibitor design, and fast protein dynamics.
We aim to understand how the diversity of animal form develops and evolved. The focus of our research is the connection between the content and organisation of genomes to the evolution of development (evo-devo). The homeobox-containing genes of the Hox gene cluster have been a corner-stone of Evolutionary Developmental Biology, but much about cluster organisation and mode of operation remains unknown.
Our aim is to gain mechanistic insight into chromatin remodelling enzymes relevant to human health – to promote the generation of new translational therapies.
My group has made major discoveries in understanding the cellular and biochemical mechanisms involved in the early stages of Alzheimer’s disease (linking mitochondrial and synaptic dysfunction), where we have pioneered new models (including the first identification that cetaceans also show similar pathology) and identified potential therapeutic targets with resultant industrial collaboration for novel inhibitors (filed patents).
Our research aims to understand how people, and other animals, defend themselves against viral infections and how viruses are able to overcome these defences. This knowledge can then inform the development of therapeutics.
We study the evolution and ecology of insect behaviour, particularly sexual and reproductive behaviour.
Our group investigates the structure and function of nano-channels connecting plant cells (plasmodesmata), and how these are hijacked by plant viruses to spread through the host.
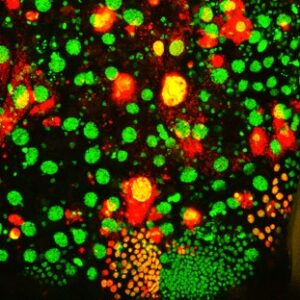
We use molecular biology, biochemistry, structural biology, live cell and super-resolution imaging, electron microscopy, reverse and forward genetics, and transient and stable plant transformation to address these questions.
We study the maturation of snRNP splicing factors and the transport of mature mRNA, with a particular emphasis on their dynamics within the nucleus and differences between neural and non-neural cell types that may be significant for SMA.
The group works on applying and developing EPR spectroscopy for studying the structure of proteins and nucleic acids. The technique uses magnetic resonance to measure the interaction between unpaired electrons and analysis reveals nanometre distances and distance distributions. The method requires that proteins and nucleic acids are tagged with the unpaired electrons.
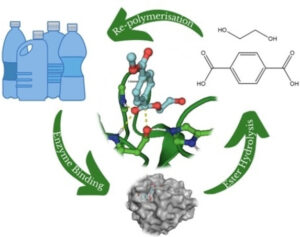
We are an Informatics and Computational Chemistry research group. Our group is based in the Purdie building on the North Haugh in St Andrews. Research areas we are interested in are machine learning, aqueous solubility, enzyme catalysis, protein-ligand interactions, modelling epidemics, molecular evolution and structural bioinformatics, computational toxicology, prediction of solubility and other molecular properties, and the classification of drugs used for doping in sport.
Our research group focuses on the study of vocal communication in mammals as a way to understand the biological basis of human speech and language and how this trait evolved.
The Sediment Ecology Research Group (SERG), known popularly as “The Mud Lab” of the Scottish Oceans Institute, University of St Andrews specialises in the investigation of the ecology of aquatic systems, with emphasis on marine and estuarine coasts, shallow marine systems and salt marshes.
We are interested in studying development in a comparative evolutionary context, with particular reference to chordates. We use the cephalochordate amphioxus as our main study species due to its relative genomic and morphological simplicity, and its phylogenetic position basal to the vertebrates.
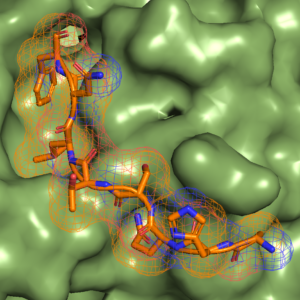
We are interested in understanding eukaryotic carbohydrate processing enzymes from a structural, mechanistic and functional viewpoint using a combination of techniques including molecular biology, protein expression, X-ray crystallography, enzyme kinetics, inhibitor design/development and cell biology.
We are interested in understanding biosynthetic processes at the molecular level and combining the tools of organic synthesis, molecular biology and protein biochemistry to generate new natural products of our own design.
Our work is highly interdisciplinary, lying on the interface of evolutionary biology, animal behaviour, ecology and psychology, and combines a wide variety of empirical and theoretical approaches.
Research in the MacNeill lab is primarily focused on dissecting the molecular biology of DNA replication and genome stability using two contrasting genetically-tractable model systems, the eukaryotic fission yeast Schizosaccharomyces pombe and the halophilic euryarchaeon Haloferax volcanii.
Prof Terry Smith is an expert in phospholipid metabolism and the use of mass spectrometry techniques and heads the Lipidomics Facility with GC-MSs and ABSciex 4000 QTrap for lipidomic and focussed metabolomics.
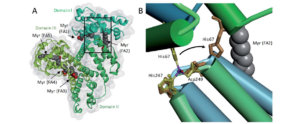
We use structural biology (NMR, X-ray crystallography, cryo-EM) combined with cell-based assays, biophysical and biochemical methods to investigate the molecular basis for interactions between pathogens and hosts. We focus on surface proteins of Gram-positive bacteria and interferon antagonists of negative-strand RNA viruses.
My research is in the field of computational biology. I examine complex biological networks, developing and evaluating algorithms to infer functional interactions based upon observational data.