Imaging Protein-Nucleic Acid Interactions Across Scales and with Single Molecule Resolution

Speaker: Prof David Rueda, Imperial College London.
Faculty of Medicine, Department of Infectious Disease
Chair in Molecular and Cellular Biophysics
Host: Prof Carlos Penedo
1st May 2024, 1 pm, BMS Seminar Room
Over the past decade, the market in RNA-based therapeutics and technology has emerged rapidly in the field of life sciences, experiencing a sizeable compound annual growth rate. Examples of early FDA-approved RNA-based drugs are spinraza, an antisense oligonucleotide developed to treat spinal muscular atrophy by regulating mRNA splicing, and patisiran, based on RNA interference to treat hereditary transthyretin-mediated amyloidosis. Other key RNA-based technologies are CRISPR/Cas9 for gene editing and, most recently, the latest RNA vaccines by Pfizer and Moderna to treat COVID-19.
To understand how RNA therapeutics function and to further develop new RNA-based drugs, it is essential to elucidate their mechanism at the molecular level. Single-molecule microscopy approaches provide unique opportunities to investigate fundamental biological processes involving nucleic acids and proteins. By eliminating ensemble-averaging, they enable monitoring such processes in real-time, providing an opportunity to track dynamic events, molecular interactions, and the formation of large macromolecular complexes and transient biomolecular species with unprecedented resolution.
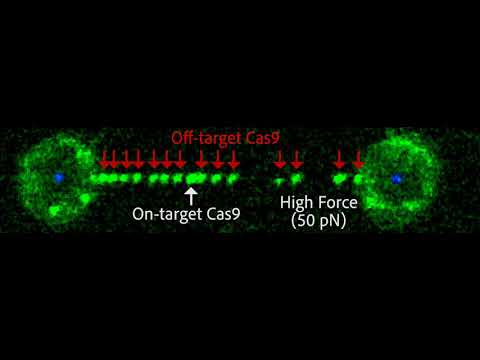
Our laboratory develops and applies fluorescence- and force-based single-molecule microscopy approaches to study fundamental biological processes involving RNA molecules and RNA-protein complexes. In this presentation, we will present our most recent efforts to determine the molecular factors that lead to CRISPR/Cas9 off-target activity and our recent advances using fluorogenic RNA aptamers for background-free imaging of cellular RNAs with single-molecule resolution.