DNA supercoiling by Atomic Force Microscopy
Base-pair resolution analysis of the effect of supercoiling on DNA structure and flexibility
Dr. Alice Pyne, University of Sheffield
28th October 2020, 1.00pm, Microsoft Teams
This is a joint seminar with the Biomedical Science Research Complex (BSRC)
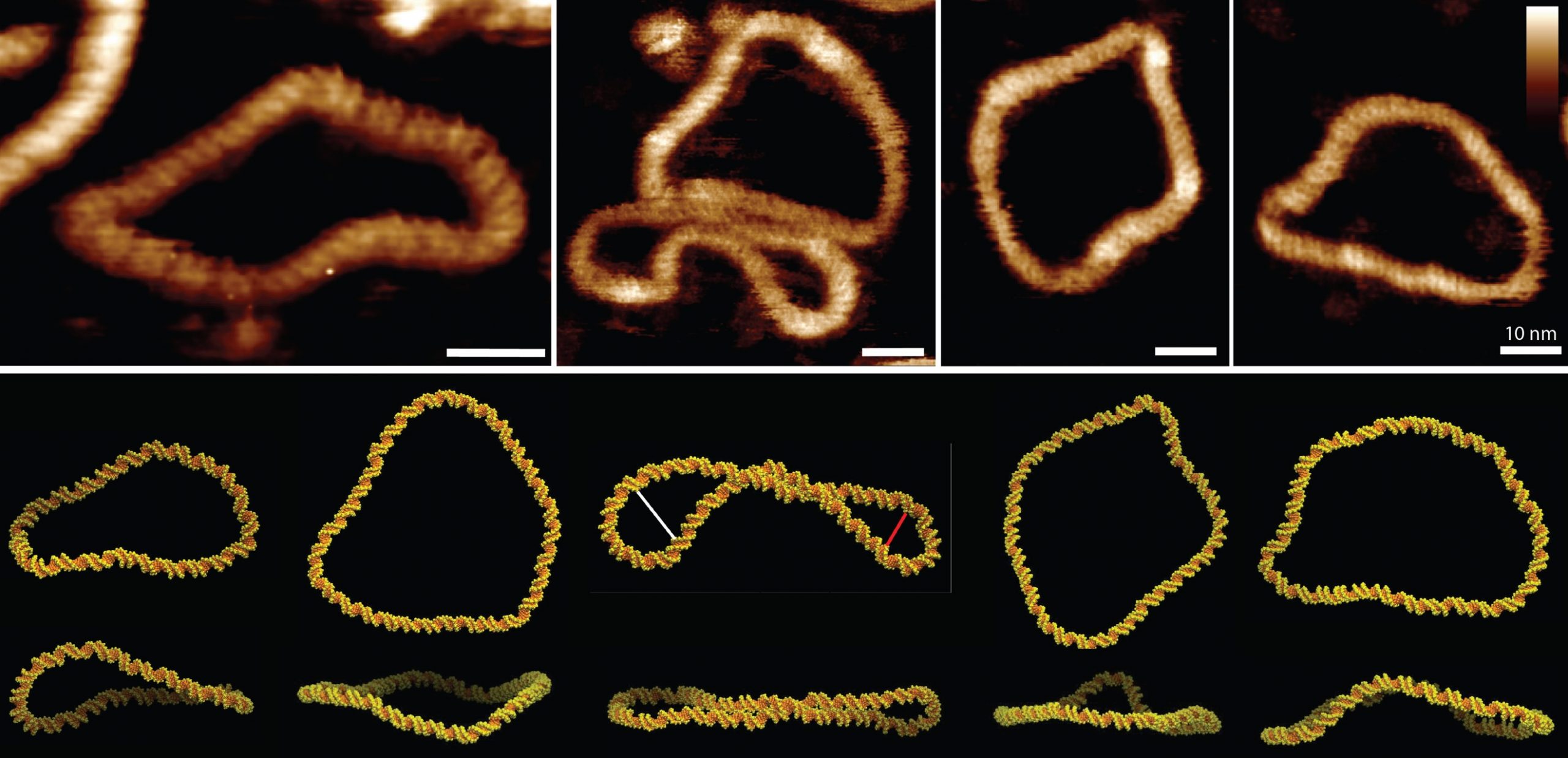
Understanding how DNA behaves in its cellular environment is a challenge of complexity, which can be enhanced by a better understanding of the fundamental properties of DNA. In the cell, DNA is arranged into highly-organised and topologically-constrained (supercoiled) structures. It remains unclear how this supercoiling affects the double-helical structure of DNA, largely because of limitations in spatial resolution of the available biophysical tools. We overcome these limitations by combining high-resolution AFM1 and atomistic MD simulations2 to resolve the structure, conformation and dynamics of supercoiled DNA to the base-pair level (Figure 1)3.
We use DNA minicircles, only twice the persistence length of DNA, to probe the structure and function of negatively-supercoiled DNA. These minicircles are small enough to be simulated at the atomistic level by MD and to be visualized at high (double-helix) resolution by AFM experiments in solution. Though the discrete, quantised nature of these minicircles implies a homogenous population, the introduction of negative supercoiling results in a large heterogenous population. We attribute this heterogeneity to the observation that negative supercoiling alone is sufficient to induce kinks and defects in the canonical B form structure of DNA. These kinks and defects affect global minicircle structure and flexibility, with DNA at this length scale appearing highly dynamic, and able to change its conformation even whilst tethered to a surface.
We propose that this dynamic conformational heterogeneity allows supercoiled DNA to bind a diverse range of substrates, and observe that both torsional and bending stress affect how DNA interacts with other oligonucleotides and proteins. We provide mechanistic insight into how DNA supercoiling can affect molecular recognition of diverse conformational substrates at sub-nanometre spatial resolution and sub second temporal resolution.
References:
- Pyne, A., Thompson, R., Leung, C., Roy, D. & Hoogenboom, B. W. Single-Molecule Reconstruction of Oligonucleotide Secondary Structure by Atomic Force Microscopy. Small 10, 3257–3261 (2014).
- Irobalieva, R. N. et al. Structural diversity of supercoiled DNA. Nature Communications 6, 8440 (2015).
- Pyne, A. L. B. et al. Base-pair resolution analysis of the effect of supercoiling on DNA flexibility and recognition. bioRxiv 863423 (2020) doi:10.1101/863423.